ISSN Print: 2278-1668, Online: 2278-0513
Some studies indicate that Janus kinase 3 (JAK3) has been involved in several types of cancer. To treat this clinical pathology, some JAK3 inhibitors have been used, such as decernotinib, and facitinib; however, some of these drugs can produce increases in liver transaminase and lipid levels. It is noteworthy that novel medications have been created to inhibit the growth of cancer cells through various experimental and theoretical approaches. Accordingly, the purpose of this study was to ascertain whether carbazole analogs (1-25) could theoretically couple with JAK3, utilizing 3pjc protein, decernotinib, and facitinib as controls within DockingServer software. Data indicate that carbazole analogs could interact at different sites of 3pxb protein surface compared to decernotinib, and facitinib. Other results suggest that inhibition constant (Ki) related to carbazole-protein complexes formation for compounds 2, 5, 9, 17, 18, and 22 was lower compared to decernotinib and facitinib drugs. This phenomenon suggests that carbazole analogs 2, 5, 9, 17, 18, and 22 could act as JAK3 blockers agents resulting in a possible decrease of cancer cell growth.
There are several data which indicate that cancer has increased in recent years worldwide,[1, 2] causing a decrease in life expectancy population.[3, 4] Several risk factors have been observed for cancer development such as alcoholism,[5] obesity,[6, 7] cigarette smoking,[8] and foods rich in fat.[9, 10] Furthermore, a study indicates that different genetic factors may be involved in different types of cancer cells;[11-16] for example, a report displayed that mutations in the KRAS gene (Kirsten rat sarcoma virus) are related to cancer cell growth.[17] Another study conducted in 436 French people with cancer indicates that 348 patients showed an established deleterious mutation in MSH2 (tumor suppressor gene).[18] Besides, a report suggests that BCR-ABL gene mutations (which encode for a tyrosine kinase) are linked to leukemia developed.[19] Another study indicates that mutations involved in HER2 (a membrane tyrosine kinase) are associated with breast cancer.[20, 21] Furthermore, a study indicates that overexpression of cMYC (regulator gene that codes for transcription factors) is associated with breast cancer.[22] Another report indicates that mutations in the EGFR gene are related to lung cancer cell growth from 15 to 20% which 10% correspond to kinase domain insertions in exon 20.[23]
Figure 1 depicts the structure of twenty-six carbazole derivatives, which were utilized to ascertain if they may interact in the following ways with the JAK3 surface:
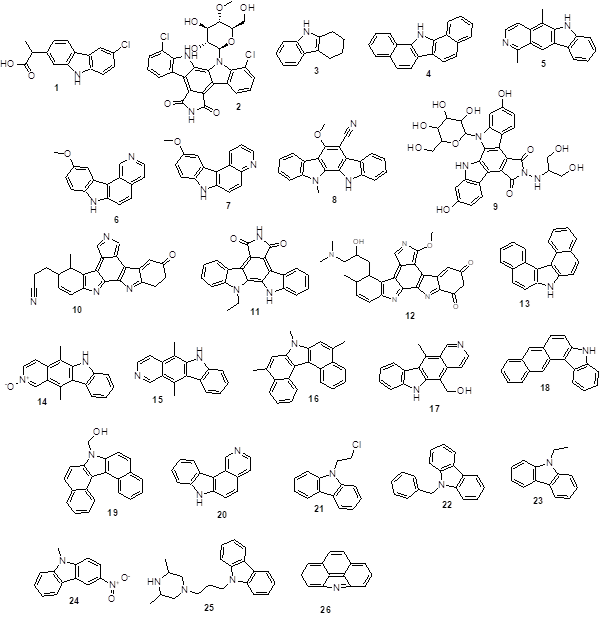 |
|
Figure 1. Chemical structure of carbazole analogs (1-26). Source: https://pubchem.ncbi.nlm.nih.gob 1 = 2-(6-chloro-9H-carbazol-2-yl)propanoic acid 2 = 5,21-dichloro-3-[(2R,3R,4R,5S,6R)-3,4-dihydroxy-6-(hydroxymethyl)-5-methoxyoxan-2-yl]-3,13,23-triazahexacyclo[14.7.0.02,10.04,9.011,15.017, 22]tricosa-1,4(9),5,7,10,15,17 (22),18,20-nonaene-12,14-dione 3 = 1,2,3,4-Tetrahydrocarbazole 4 = 1,2,7,8-Dibenzcarbazole 5 = 1,5-Dimethyl-6H-pyrido(4,3-b)carbazole 6 = 10-methoxy-7H-pyrido[4,3-c]carbazole 7 = 10-Methoxy-7H-pyrido(2,3-c)carbazole 8 = 11,12-Dihydro-6-methoxy-11-methylindolo(2,3-a)carbazole-5-carbonitri- le 9 = 6,20-dihydroxy-13-[[2-hydroxy-1-(hydroxymethyl)ethyl]amino]-3-[3,4, 5-trihydroxy-6-(hydroxymethyl)tetrahydropyran-2-yl]-3,13,23-triazahexacy-clo[14.7.0.02,10.04,9.011,15.017,22]tricosa-1(16),2(10),4,6,8,11(15),17(22),18,20-nonaene-12,14-dione 10. = 12-(2-Cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo- (2,3-a)pyrrolo(3, 4-c)-carbazole 11 = 3-ethyl-3,13,23-triazahexacyclo[14.7.0.02,10.04,9.011,15.017,22]trico-sa-1,4,6,8,10,15,17,19,21-nona-ene-12,14-dione 12 = 13-(3-dimethylamino-2-hydroxypropyl)-3-methoxy-12-methyl-6,7,12, 13-tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole-5,7-dione 13 = 3,4,5,6-Dibenzcarbazole 14 = 5,11-Dimethyl-6H-pyrido(4,3-b)carbazole 2-oxide 15 = 5,11-Dimethyl-6H-pyrido(4,3-b)carbazole 16 = 5,7,9-Trimethyl-7H-dibenzo(c,g)carbazole 17 = 5-(Hydroxymethyl)-11-methyl-6H-pyrido(4,3-b)carbazole 18 = 5H-Naphtho[2,3-c]carbazole 19 = 7-Hydroxymethyldibenzo(c,g)carbazole 20. 7H-Pyrido(4,3-c)carbazole 21 = 9-(.beta.-Chloroethyl)carbazole 22 = 9-Benzyl-9H-carbazole 23 = 9-Ethyl-9H-carbazole 24 = 9-Methyl-3-nitro-9H-carbazole 25 = 9-[3-(3,5-Dimethyl-piperazin-1-yl)-propyl]-9H-carbazole 26 = 4H-Benzo(def)carbazole |
Interaction of twenty-six carbazole derivatives with JAK3 was evaluated using 3pjc protein,[32] decernotinib and facitinib as controls in DockingServer program.[33]
Different pharmacokinetic factors for carbazole analogs (2, 5, 9, 17, 18, and 22) were determined with the use of the SwissADME software.[34]
The GUSAR software was utilized to assess the potential toxicity of carbazole analogs (2, 5, 9, 17, 18, and 22) as well as decernotinib and facitinib when administered through various routes.[35]
Certain observations in the literature suggest that analogs of carbazoles may cause changes in the development of cancer cells; for example, alectinib drug has been used as monotherapy to treat lung cancer.[36] Another study showed that compound 3,6-di(2-furyl)-9H-carbazole can act as a carcinogenic agent through topoisomerase II inhibition using MCF-7 (breast cancer) cell lines.[37] Furthermore, a report displayed that some N-acylated carbazole derivatives can decrease breast cancer cell growth using MDA-MB-231 cell lines.[38] Another report showed that a tetrahydrocarbazole derivative exerts anticancer activity against lung cancer using a Calu-1 cell line.[39] Besides, a study indicates that the compound MHY407 can produce biological activity in breast cancer using an MCF-7 cell line.[40] All of these studies suggest that some drugs are used to control the growth of cancer cells; however, their interaction with JAK3 in cancer cell growth is unclear. Thus, this study aimed to characterize the interaction of twenty-six carbazole analogs with JAK3 using 3pjc protein, decrnotinib, and facitinib as theoretical tools in the DockingServer program.
There are several protocols to characterize the protein-ligand complex formation; for example, a study displays the interaction of a carbazole-acetate derivative with tyrosinase and human glutathione receptors using the Autodock program.[41] Other studies suggest that some carbazole analogs can interact with peroxisome proliferator-activated receptor gamma using GROMAC software.[42] Besides, a report indicates that a carbazole-chrome-carboxamide analog can interact with cyclin-dependent kinase 2 using the Biovia Discovery Studio program.[43] These theoretical data suggest that some carbazoles interact with different biomolecules; therefore, the objective of this research was to characterize the coupling of twenty-six carbazoles with JAK3 using DockingServer software.[33] Table 1 and Figure 2 displayed the possible interaction of carbazole derivatives with 3pjc protein through coupling with different amino acid residues. In this way, the results indicate that Leu828 could interact with carbazole analogs 1, 3-21, and 24 in a similar manner to decrnotinib and facitinib; these data suggest that possible biological activity produced by carbazole analogs could be conditioned by the interaction with different types of amino acids involved in carbazole-JAK3 complex formation. However, it is crucial to note that the formation of carbazole-protein complexes may be influenced by certain thermodynamic parameters.
Table 1. Coupling of carbazole derivatives (1-26), decernotinib, and facitinib with amino acid residues of 3pjc protein surface. |
|
|
Compound |
Aminoacid residues |
|
Decernotinib |
Leu828; Gly829; Phe833; Lys855; Met902; Cys909; Asp912; Arg953; Asn954; Tyr904; Leu956; Asp967 |
|
Facitinib |
Leu828; Phe833; Val836; Ala853; Lys855; Glu871; Leu875; Leu900; Met902; Leu956 |
|
1 |
Leu828; Ala853; Tyr904; Leu905; Leu956 |
|
2 |
Phe833; Tyr904; Leu905; Arg911; Asp912; Leu956 |
|
3 |
Leu828; Ala853; Val884; Met902; Glu903; Tyr904; Leu956; Ala966 |
|
4 |
Leu828; Phe833; Ala853; Lys855; Glu871; Val884; Met902; Tyr904; Leu956; Asp967 |
|
5 |
Leu828; Phe833; Val836; Ala853; Lys855; Glu871; Leu875; Val884; Met902; Tyr904; Leu956; Asp967 |
|
6 |
Leu828; Phe833; Ala853; Lys855; Glu871; Val884; Met902; Tyr904; Leu956; Asp967 |
|
7 |
Leu828; Phe833; Val836; Ala853; Lys855; Glu871; Val884; Met902; Leu956; Asp967 |
|
8 |
Leu828; Phe833; Val836; Ala853; Lys855; Val884; Met902; Glu903; Tyr904; Leu956; Asp967 |
|
9 |
Leu828; Phe833; Tyr904; Arg911; Asp912; Arg953; Asn954; Leu956; Ala966; Asp967 |
|
10 |
Leu828; Phe833; Val836; Ala853; Lys855; Glu871; Val884; Leu900; Met902; Glu903; Tyr904; Leu905; Arg953; Asn954; Leu956; Asp967 |
|
11 |
Leu828; Phe833; Val836; Ala853; Lys855; Glu871; Val884; Met902; Glu903; Tyr904; Leu905; Leu956; Asp967 |
|
12 |
Leu828; Phe833; Val836; Ala853; Glu871; Leu875; Val884; Leu900; Met902; Glu903; Tyr904; Leu905; Cys909; Leu956; Al966; Asp967; Phe968 |
|
13 |
Leu828; Val836; Ala853; Val884; Met902; Glu903; Tyr904; Leu956 |
|
14 |
Leu828; Ala853; Lys855; Glu871; Val884; Met902; Tyr904; Leu956; Asp967 |
|
15 |
Leu828; Phe833; Lys855; Glu871; Leu875; Val884; Met902; Leu956; Asp967 |
|
16 |
Leu828; Phe833; Lys855; Glu871; Leu875; Val884; Met902; Leu956; Asp967 |
|
17 |
Leu828; Phe833; Val836; Ala853; Lys855; Glu871; Val884; Met902; Glu903; Tyr904; Leu956; Asp967 |
|
18 |
Leu828; Ala853; Lys855; Glu871; Leu875; Leu900; Met902; Tyr904; Leu956; Asp967 |
|
19 |
Leu828; Phe833; Val836; Ala853; Lys855; Glu871; Met902; Glu903; Asp967 |
|
20 |
Leu828; Lys855; Glu871; Leu900; Arg953; Asn954; Leu956; Ala966; Asp967 |
|
21 |
Leu828; Ala853; Lys855; Glu871; Val884; Leu900; Met902; Leu956; Asp967 |
|
22 |
Phe833; Ala853; Glu871; Leu875; Val884; Leu900; Met902; Asn954; Leu956; Ala966; Asp967 |
|
23 |
Phe833; Val836; Ala853; Lys855; Glu871; Leu875; Val884; Met902; Leu956; Asp967; Phe968 |
|
24 |
Leu828; Phe833; Val836; Ala853; Glu871; Leu875; Val884; Met902; Tyr904; Leu956 |
|
25 |
Phe833; Val836; Ala853; Val884; Met902; Cys909; Arg953; asn954; Leu956; Ala966; Asp967 |
|
26 |
Phe833; Val836; Lys855; Glu871; Leu875; Val884; Leu900; Leu956; Ala966; Asp967 |
In the literature, there are some protocols to predict Protein-ligand complex formation.[33] This research used the DockingServer program to characterize some thermodynamic parameters that can influence the formation of the carbazole-protein complex. Table 2 shows variations in energy levels for carbazole analogs compared to both decernotinib and facitinib drugs. Furthermore, the inhibition constant (Ki) was lower for carbazole analogs 2, 5, 9, 17, 18, and 22 compared with either decernotinib or facitinib drugs. These data suggest that carbazole derivatives 2, 5, 9, 17, 18, and 22 may be good JAK3 inhibitor agents. However, to confirm these findings, more research needs to be carried out using some biological models.
Table 2. Various energies at which carbazole analogs (1-26), decernotinib, and facitinib bind to the 3pjc protein surface. |
||||||
|
Compound |
A |
B |
C |
D |
E |
F |
|
Decernotinib |
-7.49 |
3.22 |
-8.24 |
0.03 |
-8.22 |
|
|
Facitinib |
-7.69 |
2.30 |
-8.60 |
-0.09 |
-8.69 |
721.73 |
|
1 |
-6.85 |
13.41 |
-7.40 |
0.16 |
-7.24 |
605.29 |
|
2 |
-8.14 |
1.09 |
-8.17 |
-0.09 |
-8.26 |
920.84 |
|
3 |
-5.88 |
48.59 |
-5.87 |
-0.02 |
-5.88 |
517.31 |
|
4 |
-8.90 |
298.71 |
-8.90 |
-0.01 |
-8.90 |
655.44 |
|
5 |
-8.12 |
1.12 |
-8.17 |
0.05 |
-8.12 |
619.48 |
|
6 |
-6.81 |
10.21 |
-7.10 |
-0.01 |
-7.11 |
610.86 |
|
7 |
-6.96 |
7.87 |
-7.29 |
0.02 |
-7.26 |
618.32 |
|
8 |
-8.29 |
840.37 |
-8.64 |
-0.03 |
-8.68 |
757.91 |
|
9 |
-8.12 |
1.12 |
-7.07 |
-0.06 |
-7.13 |
978.78 |
|
10 |
-10.0 |
40.29 |
-10.8 |
-0.10 |
-10.9 |
832.21 |
|
11 |
-9.24 |
167.36 |
-9.49 |
-0.05 |
-9.54 |
750.89 |
|
12 |
-11.2 |
5.66 |
-11.0 |
-0.16 |
-11.2 |
917.10 |
|
13 |
-9.04 |
236.64 |
-9.02 |
-0.02 |
-9.04 |
663.47 |
|
14 |
-7.52 |
4.84 |
-7.35 |
0.09 |
-7.25 |
644.57 |
|
15 |
-9.62 |
88.48 |
-9.60 |
-0.02 |
-9.62 |
789.57 |
|
16 |
-9.62 |
88.48 |
-9.60 |
-0.02 |
-9.62 |
769.57 |
|
17 |
-7.99 |
1.38 |
-7.97 |
-0.04 |
-8.02 |
635.47 |
|
18 |
-8,17 |
1.02 |
-8.17 |
0.00 |
-8.17 |
681.72 |
|
19 |
-8.72 |
405.22 |
-9.27 |
-0.05 |
-9.32 |
694.50 |
|
20 |
-6.81 |
10.21 |
-6.73 |
-0.08 |
-6.81 |
585.75 |
|
21 |
-6.59 |
14.76 |
-7.13 |
-0.02 |
-7.15 |
603.83 |
|
22 |
-7.85 |
1.75 |
-8.40 |
-0.01 |
-8.41 |
659.47 |
|
23 |
-5.97 |
41.95 |
-6.25 |
-0.02 |
-6.27 |
536.45 |
|
24 |
-6.91 |
8.59 |
-7.21 |
0.00 |
-7.21 |
563.12 |
|
25 |
-8.82 |
341.24 |
-9.30 |
-0.24 |
-9.54 |
770.48 |
|
26 |
-6.42 |
19.63 |
-6.41 |
-0.01 |
-6.42 |
516.33 |
A = Est: Free Energy of Binding (kcal/mol); B = Est. Inhibition Constant, Ki (mM)
C = vdW + Hbond + desolv Energy (kcal/mol); D = Electrostatic Energy (kcal/mol)
E = Total Intermolec. Energy (kcal/mol); F = Interact. Surface.
|
|
|
Figure 2. Coupling carbazole derivatives (2, 5, 9, 17, 18, and 22) with the 3pjc protein surface. |
Various substances with anticancer action can have their pharmacokinetic parameters predicted and analyzed using a variety of techniques; in this way, a report displayed the pharmacokinetic properties of several carbazoles using some theoretical programs to evaluate the possibility of some of these could have biological activity orally in humans.[44] For example, the pharmacokinetic properties of compound SR13668 were determined using the system version of the data from software 10.2.[45] This data suggests that some theoretical programs can be used to predict different pharmacokinetic parameters; in this way, this study aimed to characterize several pharmacokinetic factors for carbazole analogs 2, 5, 9, 17, 18, and 22 using the SwissADME program (Table 3). The determined data showed the possibility of variations in the metabolism of carbazole analogs, involving different types of cytochrome P450 isoenzymes. In this way, compound 2 shows low absorption compared with carbazole analogs 5, 9, 17, 18, and 22. Furthermore, the compound could act as an inhibitor of CYP2C9, which could cause changes in the biological activity of other drugs that are substrates of this isoenzyme such as celecoxib and flurbiprofen (nonsteroidal anti-inflammatory drugs),[46] glibenclamide and gliclazide (antidiabetic agents).[47]
Table 3. Pharmacokinetic factors for carbazole analogs 2, 5, 9, 17, 18, and 22 |
||||||
|
Parameter |
2 |
5 |
9 |
17 |
18 |
22 |
|
GI absorption BBB permeant P-GP substrate CYP1A2 inhibitor CYP2C19 inhibitor CYP2C9 inhibitor CYP2D6 inhibitor CYP3A4 inhibitor Consensus LogPO/W |
Low No No No No Yes No No 2.75 |
High Yes Yes Yes Yes No Yes Yes 3.84 |
Low No No No No No No No -0.77 |
High Yes Yes Yes Yes No Yes Yes 3.14 |
High No Yes Yes No No No No 4.82 |
High Yes Yes Yes Yes No Yes Yes 4.48 |
Some data suggest that several carbazole analogs may produce toxicity in different biological models;[48, 49] for example, a report showed that carbazol‑oxadiazol‑one analog can produce a higher toxicity degree compared with a carbazol‑acetyl derivative.[42] Analyzing these data, in this investigation the toxicity degree of carbazole derivatives 2, 5, 9, 17, 18, and 22 was determined using the Gussar program. The results indicate that compound 9 requires a higher dose through the intraperitoneal route compared to carbazole analogs 2, 5, 17, 18, and 22. These data suggest that the level of toxicity is associated with both dose and administration routes.
Table 4. Toxicity degree for carbazole analogs 2, 5, 9, 17, 18, and 20. |
||||
|
Compound |
IP LD50 (mg/kg) |
IV LD50 (mg/kg) |
Oral LD50 (mg/kg) |
SC LD50 (mg/kg) |
|
2 |
552.70 |
558.60 |
5621.00 |
3130.00 |
|
5 |
429.10 |
67.01 |
649.30 |
190.20 |
|
9 |
871.00 |
719.20 |
3459.00 |
2179.00 |
|
17 |
800.10 |
76.74 |
1278.00 |
208.70 |
|
18 |
305.20 |
72.81 |
1328.00 |
217.60 |
|
22 |
355.80 |
45.41 |
2194.00 |
723.50 |
IP - Intraperitoneal route of administration
IV - Intravenous route of administration
Oral - Oral route of administration
SC - Subcutaneous route of administration
This research has reported the interaction of carbazole derivatives to the JAK3 surface using 3pjc protein as a theoretical tool. The results suggest that carbazole derivatives 2, 5, 9, 17, 18, and 22 could have a higher affinity for 3pjc protein surface compared with the compounds 1, 3, 4-6, 6-8, 10-16, 19, 20, and 23-26. These data suggest that carbazole analogs 2, 5, 9, 17, 18, and 22 can act as JAK3 antagonists; this phenomenon could reduce cancer cell growth, which translates as a therapeutic alternative to treat some cancers.
None.
None.
None.
All procedures in this study were performed in accordance with protocols for the Pharmacochemistry Laboratory of the University Autonomous of Campeche.
|
||||||||