ISSN Print: 2278-1668, Online: 2278-0513
Several studies indicate that breast cancer has been associated with increases in mortality rates worldwide. There are several risk factors to developed breast cancer such as obesity, high fat diet, Lack of physical activity, alcohol, use of oral contraceptives, genetic mutations, and others. In addition, some reports suggesting that breast cancer gene 1 (BRCA1) has been related to breast cancer development. This research aimed to determine the possible interaction of thiophene (1) and their analogs (2 to 25) with BRCA-1 using the 3pxb protein and niraparib drug as controls in a docking model The results showed differences in the interaction of thiophene and its analogs with the surface of the 3pxb protein compared to niraparib drug. Besides, other data indicate that the inhibition constant (Ki) associated with thiophene-derivatives-protein complex formation for 11, 13, 16, 18, and 20 was similar manner to Ki for niraparib. These data suggest that compounds 11, 13, 16, 18, and 20 could inhibit the biological activity of BRCA-1; this phenomenon can be translated as a decrease in breast cancer cell growth.
For several years, breast cancer has increased drastically in women, which has increased the mortality rate worldwide.[1, 2] Some risk factors involved in the development of breast cancer have been detected, such as demographic, reproductive, hormonal, hereditary, and lifestyle parameters.[3] It should be noted that different drugs have been used to treat breast cancer such as tamoxifen (estrogen-receptor inhibitor),[4] trastuzu-mab,[5] pertuzumab,[6] margetuximab,[7] lapatinib,[8] neratinib/fulvestrant,[9] tucati-nib,[10] everolimus,[11] alpelisib,[12] capiva-sertib/fulvestrant,[13] and olaparib.[14] Nevertheless, some drugs produce some adverse effects such as nausea, diarrhea, and neutropenia.[15] In the search for new pharmacological alternatives, several drugs have been developed to treat breast cancer; for example, the preparation of niraparib (MK-4827) as an ADP-ribose polymerase inhibitory agent which is effective in BRCA-1 and BRCA-2 mutant tumors.[16] In addition, types of drugs such as olaparib and talazoparib were approved to treat breast cancers that express either BRCA-1 or BRCA-2 mutations.[17] Furthermore, a study in a phase III clinical trial showed that both olaparib and talazoparib drugs can produce beneficial effects in patients with breast cancer.[18] Other data showed that a benzo[a]pyrene and its epoxide diol can produce changes in BRCA-1 expression.[19] Furthermore, a study indicates that a carboxamide derivative has activity in a breast cancer cell line (MDA-MB-436; BRCA-1 mutated).[20]
On the other hand, some thiophene derivatives were synthesized to evaluate their biological activity against breast cancer; in this way, a study showed the preparation of a chloro-benzothiophene analog with biological activity against a breast cancer cell line (MCF-7).[21] Other report indicate that a thiophene derivative (4-Methyl-5-(phenyldiazenyl)-2-[((1-(thio- phen-2-yl) ethylidene)hydrazineylidene]- thiazol-3(2H)amine) decrease growth cancer using MCF-7 cells.[22] Other data displayed that a thiophene analog (pyrimido-thieno-pyrimidine derivative) decreases cancer cell line growth MCF-7 and A549 through epidermal growth factor receptor inhibition.[23] Besides, a dioxo-benzo[b]-thiophene derivative was developed as an agent YAP-TEAD (transcript-tional regulators) inhibitor for treating breast cancer using a cancer cell model.[24] Another report showed that a
thiophene-triazine derivative decreased the growth of MCF-7 cancer cells through PI3K/mTOR (phosphoinositide 3-kinase/mammalian target of rapamycin) pathway inhibition.[25] All these experimental data indicate that some thiophene derivatives can inhibit breast cancer growth; nevertheless, its coupling with some biomolecules involved in cell growth in breast cancer patients is unclear. Therefore, this study aimed to determine the possible interaction of several thiophene (compound 1) and their analogs (2 to 25) with BRCA-1 (breast cancer gene 1) using a theoretical model.
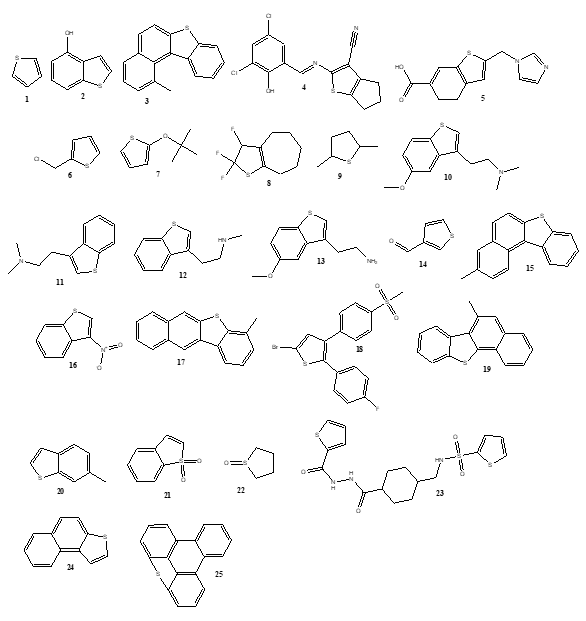 |
|
Figure 1. Chemical structure of thiophene (1) and its derivatives (2-25). 1 = Thiophene 2 = 1-Benzothiophene-4-ol 3 = 1-Methylbenzo(b)naphtho(1,2-d)-thiophene 4= 2-([(E)-(3,5-Dichloro-2-hydroxyphenyl)methylidene]amino)-5,6-dihydro-4H-cyclopenta[b]thiophene-3-carbonitrile 5 = 2-(1H-Imidazol-1-ylmethyl)-4,5-dihydrobenzo(b)thiophene-6-carboxylic acid 6 = 2-(Chloromethyl)-thiophene 7 = 2-(tert-butoxy)thiophene 8 = 2,2,3-Trifluoro-2,4,5,6,7,8-hexahydrocyclohepta(b)thiophene 9 = 2,5-Dimethyltetrahydrothiophene 10 = 3-(2-(Dimethylamino)ethyl)-5-methoxybenzo(b)thiophene 11 = 3-(2-(Dimethylamino)ethyl)benzo(b)thiophene 12 = 3-(2-(Methylamino)ethyl)benzo(b)thiophene 13 = 3-(2-Aminoethyl)-5-methoxybenzo(b)thiophene 14 = 3-Formylthiophene 15 = 3-Methylbenzo(b)naphtho(1,2-d)thiophene 16 = 3-Nitrobenzo(b)thiophene 17 = 4-Methylbenzo(b)naphtho(2,3-d)thiophene 18 = 5-Bromo-2-(4-fluoro-phenyl)-3-(4-methanesulfonyl-phenyl)-thiophene 19 = 6-Methylbenzo(b)naphtho(2,1-d)thiophene 20 = 6-Methylbenzo(b)thiophene 21 = -Benzothiophene 1,1-dioxide 22 = Tetrahydro-thiophene-1-oxide 23= N-[[4-[(thiophene-2-carbonylamino)carbamoyl]cyclohexyl]methyl]thiophene-2-sulfonamide 24 = Naphtho(2,1-b)thiophene 25= 19-thiapentacyclo[14.2.1.05,18.06,11.012,17]nonadeca-1,3,5(18), 6,8,10, 12(17),13,15-nonaene |
Coupling of thiophene and its derivatives with BRCA-1 was evaluated using 3pxb[26] protein and niraparib (MK-4827) as theoretical tools in DockingServer software.[27]
Pharmacokinetics parameter
Some pharmacokinetic parameters for thiophene derivatives (11, 13, 16, 18, and 20) were evaluated using the SwissADME software.[28]
Toxicity analysis
Possible toxicity induced through different administration routes of thiophene derivatives (11, 13, 16, 18, and 20) and niraparib was evaluated using the GUSAR program.[29]
Results and Discussion
Some reports indicate that several thiophene derivatives can exert biological activity against cancer cells;[21-25] nevertheless, experimental results are very confusing. Analyzing these data, this study aimed to evaluate the possible interaction of some thiophene and its derivatives with some biomolecules involved in cancer cell growth such as BRCA-1 using 3pxb protein and niraparib as theoretical tools in a Docking model. The results (Table 1 and Figure 2) showed that LJH685 interacts with different amino acid residues (Leu1679; Ile1680; Arg1699; Ala1700; Leu1701; Lys1702; Leu1705; Gln1779) involved in the 3pxb protein surface in comparison with thiophene (1) and its derivatives (2 to 25). This data indicates that the coupling of thiophene and its analogs with the 3pxb protein surface can depend on the chemical characteristic of each compound (Table 1 and Figure 2) or different thermodynamics parameters involved in the thiophene-protein complex formation.
Table 1. Theoretical interaction of thiophene (1) and its analogs (compounds 2-25) with 3pxb protein surface |
|
|
Compound |
Aminoacid residues |
|
Niraparib |
Leu1679; Ile1680; Arg1699; Ala1700; Leu1701; Lys1702; Leu1705; Gln1779 |
|
1 |
Ile1680; Leu1701; Lys1702; Leu1705; Gln1779 |
|
2 |
Glu1698; Arg1699; Val1740; Val1741 |
|
3 |
Glu1698; Arg1699; Ala1700; Asn1774; Met1775; Arg1699; Leu1839 |
|
4 |
Arg1699; Ala1700; Leu1701; Lys1702; Asn1774; Met1775 |
|
5 |
Ser1655; Ala1700; Leu1701; Lys1702; Phe1704; Asn1774; Met1775; Arg1835; Leu1839 |
|
6 |
Arg1699; Phe1704; Asn1774; Met1775; Arg1835; Leu1839 |
|
7 |
Leu1701; Phe1704; Asn1774; Met1775; Arg1835; Leu1839 |
|
8 |
Arg1699; Leu1701; Phe1704; Met1775; Leu1839 |
|
9 |
Phe1704; Asn1774; Met1775; Arg1835; Leu1839 |
|
10 |
Glu1698; Arg1699; Phe1704; Met1775 |
|
11 |
Glu1698; Ala1700; Leu1701 |
|
12 |
Glu1698; Arg1699; Phe1704; Met1775; Leu1839 |
|
13 |
Glu1698; Ala1700; Leu1701 |
|
14 |
Ile1680; Leu1701; Lys1702; Leu1705 |
|
15 |
Glu1698; Arg1699; Ala1700; Met1775; Leu1839 |
|
16 |
Arg1699; Leu1701; Phe1704; Met1775; Leu1839 |
|
17 |
Ala1700; Leu1701; Met1775; Arg1835; Leu1839 |
|
18 |
Glu1698; Arg1699; Ala1700; Val1740; Val1741; Thr1773; Asn1774; Met1775; Arg1835 |
|
19 |
Arg1699; Ala1700; Leu1701; Met1775; Leu1839 |
|
20 |
Arg1699; Leu1701; Phe1704; Asn1774; Met1775; Arg1835; Leu1839 |
|
21 |
Arg1699; Phe1704; Asn1774; Met1775; Leu1839 |
|
22 |
Ile1680; Leu1701; Lys1702; Leu1705 |
|
23 |
Ser1655; Glu1698; Arg1699; Ala1700; Leu1701; Asn1774; Met1775 |
|
24 |
Arg1699; Leu1701; Phe1704; Met1775; Leu1839 |
|
25 |
Glu1698; Arg1699; Leu1701; Met1775; Leu1839 |
|
|
|
Figure 2. Binding of thiophene analogs (11, 13, 16, 18, and 20) with the 3xpb protein surface. Visualized with GL mol viewer, DockingServer. |
There are theoretical studies that suggest that coupling of some drugs with several biomolecules may depend on different thermodynamic parameters, such as binding free energy, electrostatic energy, total intermolecular energy, Van Der Waals (vdW) + hydrogen bond (H bond) + desolvation energy.[29] For this reason, the study of these thermodynamic parameters involved in the interaction of thiophene and its analogs with the 3pxb protein surface was determined. The results (Table 2) displayed different energy levels for thiophene, and its analogs in comparison with niraparib. Furthermore, the inhibition constant (Ki) was higher for thiophene and its analogs 2-10, 12, 14, 15, 17, 19, and 21-25 compared to niraparib. However, Ki for thiophene analogs 11, 13, 16, 18, and 20 was very similar to the niraparib drug. These data suggest that thiophene derivatives 11, 13, 16, 18, and 20 could act as BRCA-1 inhibitory agents. However, it is important to mention that other types of biological experiments are required to verify this hypothesis.
Table 2. Thermodynamic parameters for thiophene-3pxb-protein complex formation |
||||||
|
Compound |
A |
B |
C |
D |
E |
F |
|
Niraparib |
-3.97 |
1.23 |
-4.45 |
0.17 |
-4.28 |
566.74 |
|
1 |
-2.69 |
10.64 |
-2.69 |
-0.01 |
-2.69 |
262.81 |
|
2 |
-3.20 |
4.52 |
-3.30 |
-0.19 |
-3.50 |
361.44 |
|
3 |
-5.71 |
65.54 |
-5.70 |
-0.01 |
-5.71 |
507.69 |
|
4 |
-5.15 |
166.84 |
-6.00 |
0.05 |
-5.95 |
639.36 |
|
5 |
-4.89 |
259.58 |
-5.42 |
-0.36 |
-5.78 |
502.05 |
|
6 |
-3.12 |
5.17 |
-3.41 |
-0.01 |
-3.42 |
324.30 |
|
7 |
-3.02 |
6.15 |
-3.70 |
0.01 |
-3.69 |
394.77 |
|
8 |
-5.29 |
132.58 |
-5.20 |
-0.09 |
-5.29 |
366.08 |
|
9 |
-3.09 |
5.39 |
-3.10 |
0.01 |
-3.09 |
312.43 |
|
10 |
-4.46 |
540.66 |
-4.50 |
-0.97 |
-5.47 |
479.82 |
|
11 |
-4.00 |
1.17 |
-3.98 |
-0.77 |
-4.76 |
410.80 |
|
12 |
-4.44 |
560.34 |
-4.11 |
-1.15 |
-5.26 |
426.31 |
|
13 |
-3.92 |
1.33 |
-3.92 |
-1.17 |
-5.09 |
434.79 |
|
14 |
-2.87 |
7.87 |
-3.12 |
-0.05 |
-3.17 |
293.08 |
|
15 |
-5.00 |
216.89 |
-4.99 |
0.00 |
-5.00 |
507.79 |
|
16 |
-3.77 |
1.74 |
-4.01 |
-0.05 |
-4.06 |
368.70 |
|
17 |
-5.09 |
186.30 |
-5.09 |
0.00 |
-5.09 |
493.21 |
|
18 |
-4.07 |
1.04 |
-5.12 |
-0.03 |
-5.14 |
559.29 |
|
19 |
-5.01 |
211.98 |
-5.01 |
0.00 |
-5.01 |
485.12 |
|
20 |
-3.72 |
1.89 |
-3.69 |
-0.02 |
-3.72 |
383.12 |
|
21 |
-3.40 |
3.22 |
3.38 |
-0.02 |
-3.40 |
372.32 |
|
22 |
-2.67 |
11.10 |
-2.65 |
0.02 |
-2.67 |
276.73 |
|
23 |
-2.30 |
20.55 |
-4.32 |
0.10 |
-4.21 |
616.39 |
|
24 |
-4.16 |
888.00 |
-4.15 |
-0.01 |
-4.15 |
402.99 |
|
25 |
-5.48 |
96.79 |
-5.47 |
0.00 |
-5.48 |
497.96 |
A = Est: Free Energy of Binding (kcal/mol)
B = Est. Inhibition Constant, Ki (mM)
C = vdW + Hbond + desolv Energy (kcal/mol)
D = Electrostatic Energy (kcal/mol)
E = Total Intermolec. Energy (kcal/mol)
F = Interact. Surface
Several methods have been used to determine some pharmacokinetic parameters to evaluate the possible biological activity of different drugs.[30-33] For this reason, this research aimed to determine some pharmacokinetic parameters that could be involved in the administration of thiophene analogs 11, 13, 16, 18, 20, and nirapib using the SwissADME program (Table 3). The results showed differences in gastrointestinal absorption degree and metabolism (which involves various types of cytochromes P450 enzymes) for thiophene 11, 13, 16, 18, and 20 compared to the niraparib drug. Possibly this process could be associated with the different chemical characteristics of each thiophene analog and differences in its lipophilicity degree.
Table 3. Theoretical analysis of some pharmacokinetic factors for thiophene analogs and niraparib drug |
||||||
|
Parameter |
Niraparib |
11 |
13 |
16 |
18 |
20 |
|
GI absorption BBB permeant P-GP substrate CYP1A2 inhibitor CYP2C19 inhibitor CYP2C9 inhibitor CYP2D6 inhibitor CYP3A4 inhibitor Consensus LogPO/W |
High Yes Yes Yes No No Yes No 2.29 |
High Yes No Yes Yes No Yes No 3.14 |
High Yes No Yes Yes No No No 2.49 |
High Yes No Yes Yes No No No 2.02 |
Low No No Yes Yes Yes No No 5.17 |
High Yes No Yes Yes No No No 3.19 |
Some reports suggest that different thiophene derivatives may produce some toxicity degree using several biological models.[34-37] This research aimed to evaluate the possible toxicity produced by thiophene analogs (11, 13, 16, 18, and 20) using the GUSAR software.[28] The results suggest (Table 4) that thiophene derivative 18 possibly requires high doses to produce toxicity (LD50) via intraperitoneal, intravenous, oral, and subcutaneous compared to the niraparib drug. Besides, compound 16 requires a higher dose through the oral route compared to niraparib. All these data indicate that the toxicity degree would depend on the dose and administration route of each thiophene analog.
Table 4. Theoretical analysis of toxicity for thiophene derivatives (11, 13, 16, 18, and 20) and niraparib drug using GUSAR Software |
||||
|
Compound |
IP LD50 (mg/kg) |
IV LD50 (mg/kg) |
Oral LD50 (mg/kg) |
SC LD50 (mg/kg) |
|
Niraparib |
423.10 |
104.50 |
366.00 |
158.60 |
|
11 |
90.10 |
31.09 |
190.60 |
150.50 |
|
13 |
189.80 |
75.37 |
1154.00 |
374.60 |
|
16 |
216.20 |
115.80 |
868.40 |
181.00 |
|
18 |
586.60 |
285.20 |
465.80 |
307.50 |
|
20 |
227.60 |
63.58 |
1232.00 |
416.20 |
IP - Intraperitoneal route of administration
IV - Intravenous route of administration
Oral - Oral route of administration
SC - Subcutaneous route of administration
Conclusion
In this study, the possible coupling of thiophene and its analogs with the 3pxb protein surface. The results indicate that thiophene derivatives 11, 13, 16, 18, and 20 may interact with some amino acid residues involved in the 3pxb protein surface. This data suggests that thiophene derivatives 11, 13, 16, 18, and 20 could act as BRCA-1 gene inhibitor agents; this phenomenon could be translated as a decrease in breast cancer cell growth. Analyzing these data these thiophene derivatives may be considered good pharmacological agents to treat breast cancer.
None.
None.
None.
All procedures in this study were performed in accordance with protocols for Pharmacochemistry Laboratory of University Autonomous of Campeche.
|
||||||||