ISSN Print: 2278-1668, Online: 2278-0513
Theoretical Evaluation of Furanone and its Derivatives for the Treatment of Cancer Through Eag-1 Inhibition
Magdalena Alvarez-Ramirez1, Lauro Figueroa-Valverde2*, FranciscoDiaz-Cedillo3, Marcela Rosas-Nexticapa1, Maria Lopez-Ramos2, Virginia Mateu-Armand1, Lopez-Gutierrez Tomas2
1Nutrition Laboratory, Faculty of Nutrition, University of Veracruz, Medicos y s/n Odontologos 910210, Unidad del Bosque, Xalapa, Mexico. 2Pharmacochemistry Research Laboratory, Faculty of Biological-Chemical Sciences, University Autonomous of Campeche; Humberto Lanz Cárdenas s/n, Ex Hacienda Kalá, C.P. 24085, Campeche, Mexico. 3Laboratory of Organic chemistry, Biological Sciences, National Politechnic Institute; Prolongacion de Carpio y Plan de Ayala s/n, Col. Sto Tomas, 11340, Mexico.
Abstract
Several studies suggest that some drugs such as astemizole and tetrandine can inhibit the expression of Eag-1 in cancer cells. Analyzing these data, this study aimed to evaluate the theoretical interaction of furanone (compound 1) and its derivatives (compounds 2 to 30) with Eag-1 using the 7CN1 protein as a theoretical model; in addition, astemizole, tetrandine, N-(4-(2-(Diethylamino)ethoxy)phenyl)-2-nitro-4-(trifluoromethyl)-aniline (DNTA), 1-Dimethylamino-3-[4-(2-nitro-4-trifluoromethyl-phenyl-amino)-phenoxy]-propan-2-ol (ZVS-08), and 3-Chloro-N-{2-[3,5-dibromo-4-(3-di-methyl-amino-propoxy)-phenyl]-ethyl}-4-metho-xy-benzamide (PD) were used as controls in the DockingServer software. Results showed that interaction of compounds 1-30, DNTA, ZVS-08, and PD with 7CN1 protein surface involves different aminoacid residues. Besides, inhibition constant was lower for furanone derivatives 7, 12, 16, 20, 25, 26, 29, and 30 compared to astemizole, tetrandine, DNTA, ZVS-08, and PD. These data suggest that furanone derivatives 7, 12, 16, 20, 25, 26, 29, and 30 could act as Eag-1 inhibitors in cancer cells. Therefore, these furenone derivatives could be good candidates for the treatment of cancer.
Keywords: Cancer, Furanone, Eag-1, Docking
Cancer is one of the main public health problems worldwide;[1-4] this clinical pathology can be conditioned by several factors such as alcohol,[5, 6] obesity,[4, 7] cigarette smoking,[8, 9] dietary fatty acid pattern[10, 11] and some genetic factors.[12-14] In addition, other data indicates that the Eag-1 (ether-à-go-go-1; member of the voltage-dependent potassium channel family) may be involved in cancer cell growth.[15, 16] For example, a study showed the expression of Eag-1 potassium channels in gastric cancer patients using immunohistochemistry methods.[17, 18] Another study shows that both Eag-1 mRNA and protein expression is increased in A549 lung cancer cells undergoing epithelial-to-mesenchymal transition (a likely mechanism by which tumor cells become malignant) induced by TGFβ1 (growth factor beta).[19, 20] Besides, a report displayed that Eag-1 might be involved in the pathophysiological processes of prostate cancer tissue using both reverse transcription polymerase chain reaction (RT-PCR) and immunohistochemistry (IM) methods.[21, 22] Other studies carried out in a population of 12 Chinese women with breast cancer showed that this clinical pathology was associated with Eag-1 and HIF-1α expression.[23] In addition, Eag-1 expression was determined in esophageal squamous cell carcinomas tissues through both RT-PCR and IM methods.[24, 25] To try to inhibit the cancer cells' growth some drugs such as astemizole have been used; for example, a study suggests that astemizole may decrease cervical cancer cell lines growth (HeLa, SiHa, CaSki, INBL, and C-33A) by decreasing Eag1 expression.[26-28] Other data suggest that imipramine may produce changes in Eag1 expression on a prostate cancer cell line (DU145).[29, 30] Furthermore, a study showed that calcitriol can inhibit Eag1 expression and the proliferation of human breast cancer.[31, 32] All these data suggest that some drugs can inhibit the Eag1 expression translated as a decrease in cancer cell growth; however, the interaction of these drugs with Eag1 is not very clear on cancer cells. Analyzing these data, this study aimed to evaluate the possibility that a series of furanone derivatives could interact with Eag1 using a 7CN1 protein as a theoretical model. Besides, astemizole, tetrandine, DNTA, ZVS-08, and PD drugs were used as controls on the DockingServer program.
Figure 1 shows the chemical structure of furanone and its derivatives which were used as theoretical tools to assess their potential interaction with 7CN1.
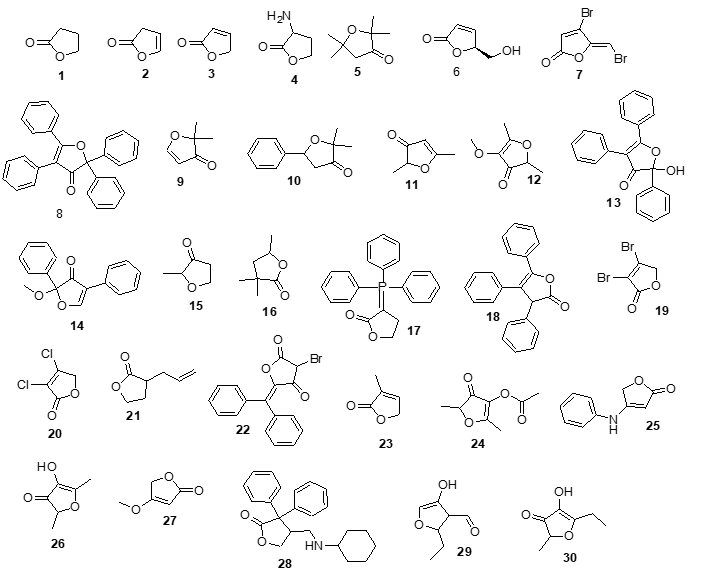 |
|
Figure 1. Chemical structure of furenone (1) and their derivatives (2-31). Source: ChemPub (https://pubchem.ncbi.nlm.nih.gov/). 1 = Dihydro-furan-2-one 2 = 3H-Furan-2-one 3 =5H-Furan-2-one 4 = Dihydro-3-amino-2-(3H)-furanone 5 = Dihydro-2,2,5,5-tetramethyl-3(2H)-furanone 6 = (S)-(-)-5-Hydroxymethyl-2(5H)-furanone 7 = (Z-)-4-Bromo-5-(bromomethylene)-2(5H)-fura-none 8 = 2,2,4,5-tetraphenyl-3(2H)-furanone 9 = 2,2-Dimethyl-3(2H)-furanone 10 = 2,2-Dimethyl-5-phenyl-dihydro-furan-3-one 11 = 2,5-Dimethyl-3(2H)-furanone 12 = 2,5-Dimethyl-4-methoxy-3(2H)-furanone 13 = 2-Hydroxy-2,4,5-triphenyl-3(2H)-furanone 14 = 2-Methoxy-2,4-diphenyl-3(2H)-furanone 15 = 2-Methyltetrahydro-3-furanone 16 = 3,3,5-trimethyldihydro-2(3H)-furanone 17 = 3-(Triphenylphosphoranylidene)dihydro-2(3H)-furanone 18 = 3,4,5-Triphenyl-2(3H)-furanone 19 = 3,4-Dibromo-2(5H)-furanone 20 = 3,4-Dichloro-2(5H)-furanone 21 = 3-Allyldihydro-2(3H)-furanone 22 = 3-Bromo-5-(Diphenylmethylene)-2(5H)-furo-none 23 = 3-Methyl-2(5H)-furanone 24 = 4-Acetoxy-2,5-dimethyl-3(2H)furanone 25 = 4-Anilino-2(5H)-furanone 26 = 4-Hydroxy-2,5-dimethyl-3(2H)-furanone 27 = 4-Methoxy-2(5H)-furanone 28 = 4-[(Cyclohexylamino)methyl]-3,3-diphenyl-dihydro-2(3H)-furanone 29 = 5-Ethyl-3-hydroxy-4-methyl-2(5H)-furenone 30 = 5-Ethyl-4-hydroxy-2-methyl-3(2H)-furanone |
The interaction of furanone and their derivatives with the Eag1 protein surface was determined using 7CN1 (PDB DOI: https://doi.org/10.2210/pdb7CN1/pdb) protein[33] as a theoretical model. In addition, to evaluate the thermodynamic parameters involved in coumarin derivative-protein complex formation, the DockingServer program was used.[34]
Theoretical pharmacokinetics involved in the chemical structure of furanone derivatives (7, 12, 16, 20, 25, 26, 29, and 30) were determined using the SwissADME software.[35]
Toxicity evaluation for furanone derivatives 7, 12, 16, 20, 25, 26, 29, and 30 was determined using GUSAR software.[36]
In the literature, there are some methods to predict the interaction of some drugs with EAG-1 such as Autodock[37] and Rosetta[38]. In this way, a study showed that tetrandrine directly interacted with Eag1 through the amino acids Ile550, Thr552, and Gln557 surface[39]. Another study showed that procyanidin B1 could interact with the EAG-1 surface which involves amino acid residues such as Ile550, Thr552, and Gln557. Analyzing these data, in this research furanone and its derivatives were used to evaluate their interaction with EAG-1 using 7CN1 protein as a theoretical model. The results shown in Table 1 indicate that the interaction of furanone and their derivatives with the 7CN1 protein surface could involve some different aminoacid-residues compared to astemizole, tetrandine, DNTA, ZVS-08, and PD drugs.
Table 1. Aminoacid residues involved in the coupling of furenone and their derivatives with 7CN1 protein surface using DockingServer |
|
|
Compounds |
Aminoacid residues |
|
Astemizole |
Pro426, Leu529, Leu532, Val533, Thr556, Leu559, Trp563, Cys566, Ile567 |
|
Tetrandine |
Leu529, Leu532, Val533, Ile560, Trp563, Ile567 |
|
DNTA |
Leu529, Leu532, Val533, Val535, leu552, Thr556, Trp563 |
|
ZVS-08 |
Pro426, Leu529, Leu532, Val533, Ala536, Trp563, Ile567 |
|
PD |
Leu532, Ile560, Trp563, Leu564, Ile567, Ile647 |
|
1 |
Asp411, Ile414, Leu415, Val418, Phe463, Arg534, Val535, Lys538, Tyr542 |
|
2 |
Asp411, Ile414, Leu415, Val418, Phe463, Arg534, Val535, Lys538, Tyr542 |
|
3 |
Asp411, Ile414, Leu415, Val418, Phe463, Arg534, Val535, Lys538, Tyr542 |
|
4 |
Ile419, , Leu552, Cys555, Thr556, Leu559 |
|
5 |
Val418, Ile419, Ala422, Leu532, Val535, Leu559 |
|
6 |
Leu415, Val418, Ile419, Ala422, Leu532, Val535, Leu552, Leu559 |
|
7 |
Leu415, Val418, Ile419, Leu532, Val535, Tyr542 Leu552, Cys555, Thr556, Leu559 |
|
8 |
Ile419, Ala422, Leu532, Val535, Ala536, Leu552, Cys555, Thr556, Leu559, Trp563 |
|
9 |
Leu415, Val418, Ile419, Val535, Tyr542, Leu552 |
|
10 |
Thr425, Pro426, Ala429, Thr526, Leu529, Trp563, Cys566, Ile567 |
|
11 |
Val418, Ile419, Ala422, Leu532, Val535, Leu552, Leu559 |
|
12 |
Leu415, Val418, Ile419, Ala422, Leu532, Val535, Leu552, Leu559 |
|
13 |
Ile419, Leu532, Val535, Ala536, Leu552, Cys55, Thr556, Leu559, Trp563 |
|
14 |
Leu415, Val418, Ile419, Val535, Tyr542, Leu552, Leu553, Thr556, Leu559 |
|
15 |
Leu415, Val418, Ile419, Val535, Tyr542, Leu552 |
|
16 |
Val418, Ile419, Ala422, Leu532, Val535, Leu552, Leu559 |
|
17 |
Ile419, Ala422, Leu532, Val535, Ala536, Leu552, Thr556, Leu559, Trp563 |
|
18 |
Leu415, Val418, Ile419, Val535, Leu539, Leu552, Cys555Thr556, Leu559, Trp563 |
|
19 |
Leu415, Val418, Ile419, Leu532, Val535 |
|
20 |
Val418, Ile419, Leu532, Val535 |
|
21 |
Leu415, Val418, Ile419, Ala422, Leu532, Val535 |
|
22 |
Pro426, Leu529, Leu532, Trp563, Ile567 |
|
23 |
Leu415, Val418, Ile419, Val535, Tyr542, Leu552 |
|
24 |
Val418, Ile419, Ala422, Leu532, Val535, Leu552, Cys555, Thr556, Leu559 |
|
25 |
Val418, Ile419, Ala422, Leu532, Leu552, Thr556, Leu559 |
|
26 |
Val418, Ile419, Leu532, Leu552, Leu559 |
|
27 |
Leu415, Val418, Ile419, Ala422, Leu532, Val535, Tyr542, Leu552 |
|
28 |
Thr425, Pro426, Ala429, Thr526, Leu529, Leu532, Trp563, Cys566, Ile567 |
|
29 |
Val418, Ile419, Ala422, Leu532, Leu552, Leu559 |
|
30 |
Leu415, Val418, Ile419, Ala422, Leu532, Val535, Leu552, Leu559 |
DNTA = N-(4-(2-(Diethylamino)ethoxy)phenyl)-2-nitro-4-(trifluoromethyl)-aniline; ZVS-08 = 1-Dimethylamino-3-[4-(2-nitro-4-trifluoromethyl-phenyl amino)-phenoxy]-propan-2-ol; PD = 3-Chloro-N-{2-[3,5-dibromo-4-(3-di-methylamino-propoxy)-phenyl]-ethyl}-4-methoxy-benzamide.
However, it is important to mention that this interaction may involve some thermodynamic parameters such as the free energy of binding, electrostatic energy, total intermolecular energy, and 4) Van der Waals (vdW) + hydrogen bond (H-bond) + desolvation energy.[34] For this reason, in this study, some thermodynamic parameters involved in the interaction of furanone and its analogs with the 7CN1 protein surface were evaluated using the DockingServer program. The results (Table 2) display differences in bond-energy levels for furanone and their derivatives compared with astemizole, tetrandibne, DNTA, ZVS-08, and PD. Besides, the inhibition constant (Ki) was lower for furonone derivatives 7, 12, 16, 20, 25, 26, 29 and 30 compared to PD. Other results indicate that Ki for furanone derivatives 1-9, 11-13, 15, 16. 18-21, 23, and 25-30 were lower compared to astemizole. All these data suggest that these furanone derivatives (Figure 2) could act as EAG-1 inhibitors, resulting in a decrease in cancer cell growth.
Table 2. Thermodynamic parameters involved in the interaction of coumarin and their derivatives with 7CN1 protein surface using DockingServer software. |
||||||
|
Compound |
A |
B |
C |
D |
E |
F |
|
Aztemisole |
-5.91 |
46.76 |
-8.29 |
0.38 |
-7.91 |
828.65 |
|
Tetrandine |
-5.21. |
151.33 |
-6.95 |
0.72 |
-6.22 |
769.28 |
|
DNTA |
-4.65 |
389.22 |
-6.19 |
0.44 |
-5.75 |
728.23 |
|
ZVS-08 |
-5.41 |
108.97 |
-6.10 |
0.44 |
-5.66 |
638.51 |
|
Purperealidin analog |
-3.57 |
2.42 |
-5.29 |
0.29 |
-4.99 |
674.35 |
|
1 |
-2.82 |
8.59 |
-3.00 |
0.18 |
2.82 |
220.51 |
|
2 |
-2.73 |
10.05 |
-2.47 |
-0.25 |
-2.73 |
217.38 |
|
3 |
-3.09 |
6.42 |
-2.75 |
-0.34 |
-3.09 |
217.50 |
|
4 |
-2.23 |
23.02 |
-2.93 |
0.39 |
-2.53 |
323.42 |
|
5 |
-4.15 |
910 |
-4.16 |
0.01 |
-4.15 |
418.33 |
|
6 |
-3.19 |
4.56 |
-2.88 |
-0.01 |
-2.89 |
321.75 |
|
7 |
-4.05 |
1.08 |
-4.04 |
0.00 |
-4.05 |
306.14 |
|
8 |
-6.48 |
17.81 |
-7.66 |
0.01 |
-7.64 |
734.20 |
|
9 |
-3.03 |
5.96 |
-3.03 |
0.00 |
-3.03 |
326.04 |
|
10 |
-5.16 |
164.40 |
-5.46 |
0.00 |
-5.46 |
473.79 |
|
11 |
-3.66 |
2.09 |
-3.20 |
-0.46 |
-3.66 |
333.45 |
|
12 |
-3.61 |
2.25 |
-3.46 |
-0.45 |
-3.91 |
418.17 |
|
13 |
-6.12 |
32.71 |
-7.01 |
0.00 |
-7.01 |
678.55 |
|
14 |
-5.56 |
83.62 |
-6.43 |
0.00 |
-6.44 |
612.84 |
|
15 |
-3.01 |
6.20 |
-3.01 |
0.00 |
-3.01 |
305.43 |
|
16 |
-3.81 |
1.60 |
-3.81 |
0.00 |
-3.81 |
354.91 |
|
17 |
-5.50 |
93.62 |
-6.89 |
0.01 |
-6.88 |
698.70 |
|
18 |
-6.47 |
18.23 |
-7.39 |
-0.01 |
-7.40 |
656.12 |
|
19 |
-3.21 |
4.45 |
-3.21 |
0.00 |
-3.21 |
277.67 |
|
20 |
-4.04 |
1.10 |
-4.04 |
0.00 |
-4.04 |
361.31 |
|
21 |
-3.42 |
3.10 |
-4.01 |
0.00 |
-4.00 |
365.16 |
|
22 |
-5.21 |
152.69 |
-5.84 |
-0.01 |
-5.85 |
512.90 |
|
23 |
-2.73 |
9.95 |
-2.74 |
-0.01 |
-2.73 |
303.10 |
|
24 |
-4.21 |
823.80 |
-4.31 |
-0.44 |
-4.75 |
453.04 |
|
25 |
-3.60 |
2.29 |
-4.20 |
0.00 |
-4.19 |
477.50 |
|
26 |
-3.66 |
2.07 |
-3.22 |
-0.44 |
-3.66 |
353.21 |
|
27 |
-2.56 |
13.24 |
-2.85 |
-0.01 |
-2.86 |
319.19 |
|
28 |
-7.14 |
5.83 |
-7.66 |
0.41 |
-7.25 |
605.97 |
|
29 |
-3.85 |
1.50 |
-3.70 |
-0.45 |
-4.15 |
403.21 |
|
30 |
-3.64 |
2.14 |
-3.49 |
-0.45 |
-3.94 |
387.40 |
A = Est: Free Energy of Binding (kcal/mol); B = Inhibition Constant, Ki (mM); C = vdW + Hbond + desolv Energy (kcal/mol); D = Electrostatic Energy (kcal/mol); E = Total Intermolec. Energy (kcal/mol); F = Interact. Surface; DNTA = N-(4-(2-(Diethylamino)- ethoxy)phenyl)-2-nitro-4-(trifluoromethyl)aniline; ZVS-08 = 1-Dimethylamino-3-[4-(2-nitro-4-trifluoromethyl-pheny-lamino)-phenoxy]-propan-2-ol.
|
|
|
|
|
|
|
|
|
|
|
|
|
Figure 2. Interaction of furanone derivatives (7, 12, 16, 20, 25, 26, 29, AND 30) with 7CNC1 protein surface using Dockingserver software |
|
Different methods have been used to predict some pharmacokinetic parameters involved in some anticancer drugs.[40, 41] Analyzing these data, in this investigation, some pharmacokinetic parameters for furanone derivatives 7, 12, 16, 20, 25, 26, 29, and 30 were evaluated using the SwissADME program.[42] Tables 3 and 4 showed the pharmacokinetic parameters involved in the possible gastrointestinal absorption and metabolism of furonone derivatives, which involve some cytochrome P450 systems. This phenomenon could depend on the chemical structure of furanone derivatives and their lipophilicity degree.
Table 3. Pharmacokinetic parameters for astemizole (i), tetrandine (ii), DNTA (iii), ZVS (iv), PD (v) and furanone derivative (7). |
||||||
|
Parameter |
i |
ii |
iii |
iv |
v |
7 |
|
GI absorption BBB permeant P-gp substrate CYP1A2 inhibitor CYP2C19 inhibitor CYP2C9 inhibitor CYP2D6 inhibitor CYP3A4 inhibitor Consensus LogPO/W |
H Yes Yes No Yes No Yes Yes 5.00 |
H No No No No No No No 5.49 |
H No No No Yes Yes Yes Yes 4.19 |
H No Yes No Yes Yes Yes Yes 3.02 |
H Yes No Yes Yes Yes Yes Yes 5.10 |
H Yes No No No No Yes No 1.81 |
H = high; i = Astemizol; ii = tetrandine; iii DNTA; iv = ZVS-08; v = PD . GI = gastrointestinal; BBB = Blood-Brain-Barrier; P-gp = P-glycoprotein.
Table 4. Pharmacokinetic parameters for furanone derivatives 12, 16, 20, 25, 26, 29 and 30. |
|||||||
|
Parameter |
12 |
16 |
20 |
25 |
26 |
29 |
30 |
|
GI absorpt BBB perm P-gp substrate CYP1A2 inhibitor CYP2C19 inhibitor CYP2C9 inhibitor CYP2D6 inhibitor CYP3A4 inhibitor Consensus LogPO/W |
H Yes No No No No No No 0.92 |
H Yes No No No No No No 1.63 |
H Yes No No No No No No 1.41 |
H Yes No Yes No No No No 1.47 |
H Yes No No No No No No 0.56 |
H Yes No No No No No No 0.56 |
H No No No No No No No 0.25 |
In the literature, there are some methods to predict the degree of toxicity of various compounds such as ADME/Tox,[43] eToxPred,[44] and GUSSAR.[45] This study aimed to evaluate the possible toxic effect produced by furanone derivatives 7, 12, 16, 20, 25, 26, 29, and 30 using the GUSSAR software. The results shown in Table 5 suggest that higher doses of furanone derivatives (7, 12, 16, 20,25, 26, and 29) are needed (via intraperitoneal) to produce toxicity compared to astemizol, tetrandine, DNTA, ZVS-08, AND PD drugs. Besides, other data indicate that furanone derivatives 7, 12, 16, 20,25, 26, 29, and 30 require higher doses (via oral) to induce toxicity compared to astemizol, tetrandine, DNTA, ZVS-08, AND PD drugs.
Table 5. Theoretical toxicity for furanone derivatives |
||||
|
Compound |
IP LD50 (mg/kg) |
IV LD50 (mg/kg) |
Oral LD50 (mg/kg) |
SC LD50 (mg/kg) |
|
Astemizole |
253.30 |
36.59 |
835.00 |
710.90 |
|
Tetrandine |
70.92 |
65.40 |
708.30 |
121.80 |
|
DNTA |
345.80 |
84.54 |
992.80 |
473.80 |
|
ZVS-08 |
286.40 |
83.93 |
727.30 |
579.20 |
|
PD |
330.10 |
80.65 |
960.00 |
4990.00 |
|
7 |
496.40 |
29.71 |
910.00 |
856.40 |
|
12 |
263.20 |
40.96 |
872.00 |
651.50 |
|
16 |
426.00 |
14.56 |
2726.00 |
951.30 |
|
20 |
726.30 |
18.92 |
785.30 |
1242.00 |
|
25 |
653.70 |
32.21 |
909.50 |
1084.00 |
|
26 |
125.90 |
42.37 |
861.10 |
565.80 |
|
29 |
385.50 |
60.90 |
1975.00 |
727.70 |
|
30 |
181.40 |
67,78 |
1322.00 |
522.80 |
IP = Intraperitoneal.
IV = Intravenous.
Oral = Oral.
SC = Subcutaneous.
In this research, the theoretical interaction of furanone and its derivatives with Eag-1 was determined. The results showed a higher affinity of furanone derivatives 7, 12, 16, 20, 25, 26, 29, and 30 for Eag-1 surface compared to astemizol, tetrandine, DNTA, ZVS-08 AND PD drugs. All these data suggest that furanone derivatives 7, 12, 16, 20, 25, 26, 29, and 30 could act as Eag-1 inhibitors. This phenomenon could be translated as good candidates for the treatment of cancer cells.
None.
None.
None.
All procedures in this study were performed in accordance with protocols for Pharmacochemistry Laboratory of University Autonomous of Campeche.
|
||||||||